Evaluating the safety and efficacy of new drugs
Testing drugs in living organisms is a necessary component of drug development. The journey of a newly discovered drug candidate begins with pre-clinical testing in molecular, cell and animal studies. At this stage, the therapeutic mechanism of the drug is defined, its safety in animal tissues determined and structural variants will be screened for maximum efficacy. Only after safety and efficacy have been proven in animals can the drug candidate proceed to clinical trials in humans. Human clinical trials are split into four sub-phases, which assess different levels of safety and efficacy and optimize dosing. Ultimately, the clinical trials must determine that the drug has a superior effect on the patient's disease compared with a placebo and that the beneficial effect outweighs any adverse effects.
The framework of evaluating a new drug is based on several incremental assumptions, if a drug is safe and effective in animals it should be safe and effective in humans, if the drug performs well in a controlled trial of humans it should perform comparably for the wider population. Nevertheless, almost 90% of drugs fail somewhere in the process, suggesting that in many cases the incremental process of pre-clinical and clinical testing is not a good predictor of success.
Working towards better predictors of success
Tissue simulation and computational modelling may hold the key to unlocking the predictive power of pre-clinical testing. Human tissue simulation technologies mimic the biological functions of real human tissues, giving the mechanistic insights made possible by in vitro experiments, as well as organ and organism scale data usually obtained through animal testing. Such technologies have the potential to significantly reduce time in pre-clinical trials and eliminate ethical concerns associated with testing on animals, by using lab-made chips that accurately simulate human organs. In silico clinical trials could be used to predict the outcomes of human clinical trials before the event and identify any
Organ-on-a-chip
Organ-on-a-chip (OOC) devices are defined as multi-channel 3-D microfluidic cell culture chips that simulate the activities, mechanics and physiological responses of human tissues. After tissue culture, OOCs represent the next level of development towards high-fidelity artificial organ systems. While tissue culture systems can mimic higher levels of cell differentiation and even tissue organization, OOCs take this one step further by incorporating dynamic properties of the tissues. Such properties include spatiotemporal gradients of chemicals and mechanical movements of an organ. OOCs exist for a number of human tissues, such as the lung, heart, kidney, artery and skin. Some progress has also been made towards connecting these tissues via thin external tubes to create a 'body-on-a-chip'.
Several OOCs were first developed in academic labs. For instance, the lung-on-a-chip came out of the Wyss Institute for Biologically Inspired Engineering at Harvard in 2010, and one of the first kidney-on-a-chips was created at MIT in 2008. Many have since been spun-out for commercial development. Currently, several companies have made OOCs commercially available, such as EmulateBio. The Boston-based start-up develops USB-stick sized OOCs for the liver, lung and intestine, with additional tissues in development. Emulate currently has active research partnerships with pharmaceutical companies, including AstraZeneca, Roche, Takeda, Merck, Janssen and Seres Therapeutics. In 2017, the FDA signed a collaborative agreement with Emulate to test their OOC models as predictors of safety for foods, dietary supplements and cosmetics. These devices are primarily positioned as an alternative to animal testing in the pharmaceutical industry, but are also in development for toxicology testing in consumer goods industries such as cosmetics or F&B.
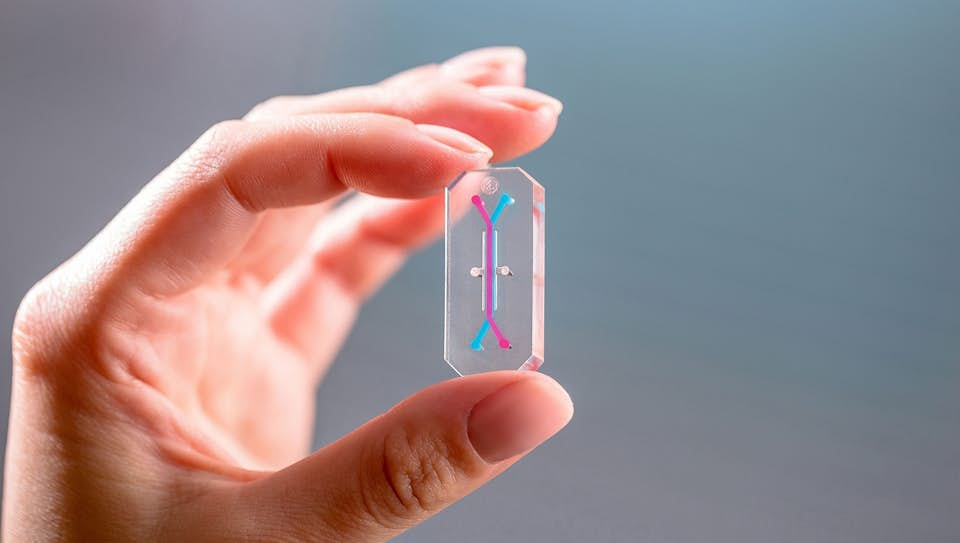
EmulateBio develops USB-stick sized OOCs for the liver, lung and intestine, with additional tissues in development
In silico clinical trials
In silico clinical trials are individualized computer simulations used in the development or regulatory evaluation of a medicinal product, device, or intervention. They have the potential to deliver significant cost and time savings in clinical trial planning, with only a small team of specialists required to operate the simulation. Clinical trials that would usually take years to plan, conduct and follow-up, could be simulated in a matter of days.
Many in silico models are created using data from the scientific literature on human physiology and in vivo clinical trial data. Biochemical pathways that have been characterized in the laboratory can be converted into mathematical equations and modelled computationally. Once the tissue function is successfully modelled, the mechanism of pharmaceutical drugs can be fed into the model to predict their primary effects on the disease state, effects on the disease progression (with a temporal capability) and their secondary effects on the rest of the tissue. Theoretically, this would allow a much higher throughput than in vitro or in vivo studies, allowing the efficacy of hundreds of drug candidates to be screened rapidly, and detailed mechanistic predictions to be made.
The virtual physiological human
The potential cost and time advantages of computer simulation has attracted numerous initiatives to develop these technologies, at the academic, commercial and governmental level. For example, the Virtual Physiological Human is a European Initiative to develop a methodological and technological framework, enabling collaborative investigation of the human body as a single complex system. The European Commission state that "the virtual physiological human will revolutionise the way health knowledge is produced, stored and managed as well as the way in which healthcare is currently delivered." The project is in its early stages with the first phase completed in September 2014, which yielded a research and technological development roadmap and strategy for in silico clinical trials. The published (non-exhaustive) list of VPH funded projects has just under 150 entries, including virtual physiological organs, disease states, diagnosis and patients.
Virtual assay
The virtual assay platform has been developed by researchers at the University of Oxford. The virtual assay is provided as a software to run in silico drug trials in populations of human cardiac cell models for predictions of drug safety and efficacy in cardiotoxicity studies. By modulating variables in the cellular models, the aim is to mimic a real population of patients, who may have subtle differences in their physiology and tissue functioning. The simulated population is screened against in vivo experimental data so that only virtual patients that can be calibrated to real-patients are retained. The calibrated virtual population can then be modelled with the presence of different pharmaceutical drugs.
This assay has been tested for its ability to predict drug-induced arrhythmias based on ion channel modelling. The simulation results were directly compared with existing experimental data for 62 reference compounds, and the virtual assay was found to predict clinical risk with 89% accuracy - a higher accuracy than animal experiments. The software is currently being evaluated in several studies with pharmaceutical companies.
Conclusions
OOCs and in silico simulation technologies have immense potential to reduce the time and costs associated with the development of new drugs, and to make the clinical trial process safer and more efficient. The technologies that are currently in development are showing promise, with rapid commercialization and up to 89% accuracy in the prediction of clinical risk. Beyond simulating clinical trials, in silico modelling technologies could also hold the key to revealing insights from swathes of data in the medical and scientific literature leading to better diagnostics, treatments and healthcare for patients.

Dr Rachel Murkett
Project Director
Dr Rachel Murkett leads strategic intelligence projects at Biochromex. She holds a PhD in Chemistry from the University of Cambridge and has 6+ years of experience as a scientific consultant to consumer goods brands.